Email:yhhe@pku.edu.cn
Fax:
Tel: 86-010-62765439

Introduction
Dr. Yuehui HE
Professor & Vice Dean, School of Advanced Agricultural Sciences, Peking University
Principal Investigator, Peking-Tsinghua Center for Life Sciences
Education
Ph.D. (2001), The University of Kentucky, USA
Working Experience
2020-now, Professor, School of Advanced Agricultural Sciences & Peking-Tsinghua Center for Life Sciences, Peking University
2014-2020, Senior Principal Investigator, Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences
2012-2014, Associate Professor (tenured), Department of Biological Sciences, National University of Singapore, Singapore
2012-2014, Senior Principal Investigator, Temasek Life Sciences Laboratory, Singapore
2006-2011, Principal Investigator, Temasek Life Sciences Laboratory, Singapore
2005-2011, Assistant Professor, Department of Biological Sciences, National University of Singapore, Singapore
2001-2005, Postdoctoral Research Associate, Department of Biochemistry, University of Wisconsin-Madison, USA
Research interests
Plant environmental epigenetics
Epigenetic mechanisms in vernalization and flowering-time regulation
Crop genetic improvement of flowering time
Research introduction
Chromatin modifications and remodeling play critical roles in chromatin-templated processes such as gene expression. The Laboratory of Plant Environmental Epigenetics explores epigenetic or chromatin-mediated mechanisms for plant-environment interactions, with a main focus on environmental regulation of the timing of developmental transition to flowering (flowering time).
When to flower is often timed by seasonal cues including day-length changes and/or winter cold to ensure that flowering occurs at a season favorable for seed production. Many over-wintering plants grown at high latitudes acquire competence to flower in spring or early summer, through a physiological process termed as vernalization. Vernalization-responsive plants often remember past winter cold experience when temperature rises in spring to enable flowering. We are interested in how epigenetic ‘memory of winter cold’ is established, maintained and subsequently erased (reset) in diverse plants, with a focus on molecular epigenetic mechanisms underlying vernalization regulation in the model plant Arabidopsis thaliana and the temperate cereal wheat (Triticum aestivum). In addition, we are interested in molecular mechanisms underlying photoperiodic induction of flowering in vernalization-responsive plants including Arabidopsis and wheat.
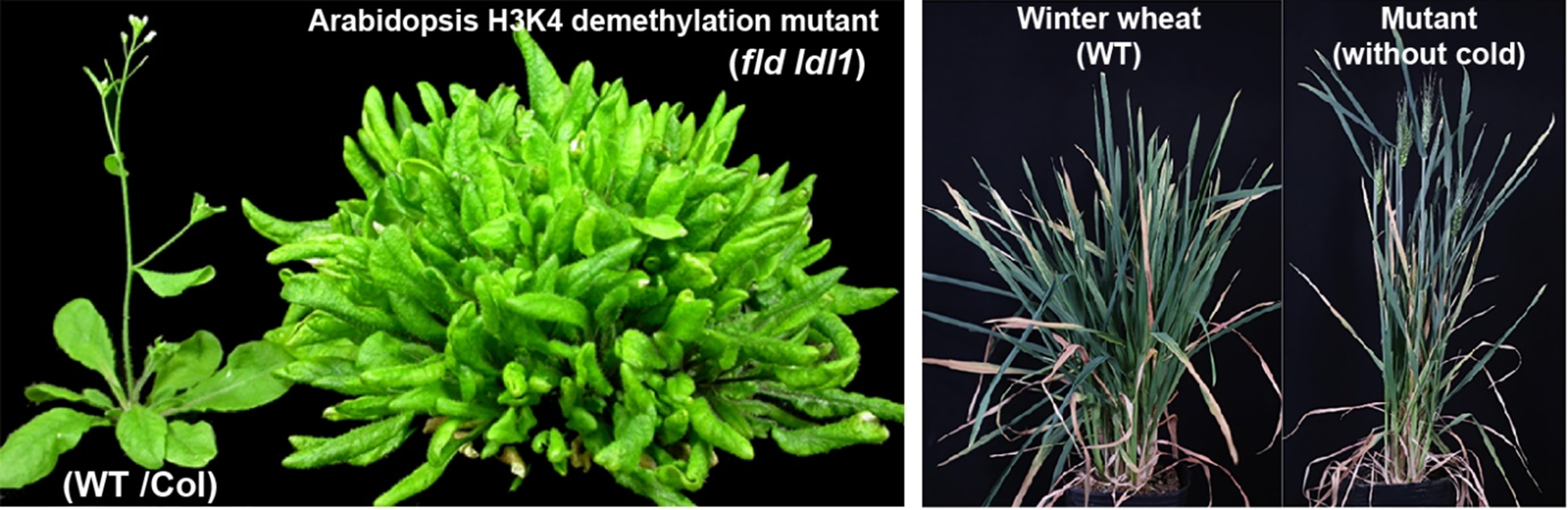
Primary cellular metabolisms have been long known to influence nuclear gene expression in plants. Over the years, we have identified a few Arabidopsis flowering-time mutants harboring defective enzymes involved in cellular primary metabolisms. We are interested in metabolic control of gene expression through chromatin-mediated mechanisms, with the aim of elucidating how metabolic flux shapes the plasticity of plant growth and development in response to environmental stimuli.
Selected Publications
Huang F & He Y. (2024) Epigenetic control of gene expression by cellular metabolisms in plants. Curr. Opin. Plant Biol, 81: 102572.
Gao Z & He Y. (2024) Molecular epigenetic understanding of winter memory in Arabidopsis. Plant Physiol, 194: 1952-1961.
Niu D, Gao Z, Cui B, Zhang Y & He Y. (2024) A molecular mechanism for embryonic resetting of winter memory and restoration of winter annual growth habit in wheat. Nature Plants, 10: 37-52.
Huang F, Luo X, Ou Y, Gao Z, Tang Q, Chu Z, Zhu X & He Y. (2023) Control of histone demethylation by nuclear-localized α-ketoglutarate dehydrogenase. Science 381: eadf8822.
Zhang Z, Luo X, Yang, Y & He Y. (2023) Cold induction of nuclear FRIGIDA condensation in Arabidopsis. Nature 7969: E27-E32.
Gao Z, Li Y, Ou Y, Yin M, Chen T, Zeng X, Li R & He Y. (2023) A pair of readers of bivalent chromatin mediate formation of Polycomb-based "memory of cold" in plants. Molecular Cell 83: 1109-1124.
Xu G, Tao Z & He Y. (2022) Embryonic reactivation of FLOWERING LOCUS C by ABSCISIC ACID-INSENSITIVE 3 establishes the vernalization requirement in each Arabidopsis generation. Plant Cell 34: 2205-2221.
Li Z, Luo X, Ou Y, Jiao H, Peng L, Fu X, Macho AP, Liu R & He Y. (2021) JASMONATE-ZIM DOMAIN proteins engage Polycomb chromatin modifiers to modulate Jasmonate signaling in Arabidopsis. Molecular Plant, 14: 732-747.
Luo X, Ou Y, Li R & He Y. (2020) Maternal transmission of the epigenetic 'memory of winter cold' in Arabidopsis. Nature Plants 6: 1211-1218.
Tao Z, Hu H, Luo X, Jia B, Du J & He Y. (2019) Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis. Nature Plants 5: 424-435.
Luo X, Chen T, Zeng X, He D & He Y. (2019) Feedback regulation of FLC by FLOWERING LOCUS T and FD through a 5' FLC promoter region. Molecular Plant 12: 285-288.
Li Z, Jiang D & He Y. (2018) FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nature Plants 4: 836-846.
Li Z, Fu X, Wang Y, Liu R & He Y. (2018) Polycomb-mediated gene silencing by the BAH-EMF1 complex in plants. Nature Genetics 50: 1254-1261.
He Y. and Li Z. (2018) Epigenetic environmental memories in plants: establishment, maintenance, and reprogramming. Trends in Genetics 34: 856-866.
Li Z, Ou Y, Zhang Z, Li J & He Y. (2018) Brassinosteroid signaling recruits Histone 3 lysine-27 demethylation activity to FLOWERING LOCUS C chromatin to inhibit the floral transition in Arabidopsis. Molecular Plant 11: 1135-1146.
Tao Z, Shen L, Gu X, Wang Y, Yu H & He Y. (2017) Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551: 124–128.
Yuan W, Luo X, Li Z, Yang W, Wang Y, Liu R, Du J & He Y. (2016) A cis cold memory element and a trans epigenome reader mediate Polycomb silencing of FLC by vernalization in Arabidopsis. Nature Genetics 48: 1527-1534.
Li Z, Jiang D, Fu X, Luo X, Liu R & He Y. (2016) Integration of histone methylations with RNA processing by the nuclear mRNA Cap Binding Complex. Nature Plants 2: 16015.
Wang Y., Gu X., Yuan W., Schmitz R. & He Y. (2014) Photoperiodic control of the floral transition through a distinct Polycomb repressive complex. Developmental Cell 28: 727-736.
Gu X, Wang Y & He Y. (2013) Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biology, 11: e1001649.
Gu X, Le C, Wang Yi, Li Z, Jiang D, Wang Yu & He Y. (2013) Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nature Communications 4 (1947): 1-10.
He Y. (2012) Chromatin regulation of flowering (invited review). Trends in Plant Science 17: 556-562.



